| |
|
||||||||||
Wichtiger Hinweis:
Diese Website wird in älteren Versionen von Netscape ohne graphische Elemente dargestellt. Die Funktionalität der Website ist aber trotzdem gewährleistet. Wenn Sie diese Website regelmässig benutzen, empfehlen wir Ihnen, auf Ihrem Computer einen aktuellen Browser zu installieren. Weitere Informationen finden Sie auf
folgender Seite.
Important Note:
The content in this site is accessible to any browser or Internet device, however, some graphics will display correctly only in the newer versions of Netscape. To get the most out of our site we suggest you upgrade to the latest Netscape.
More information
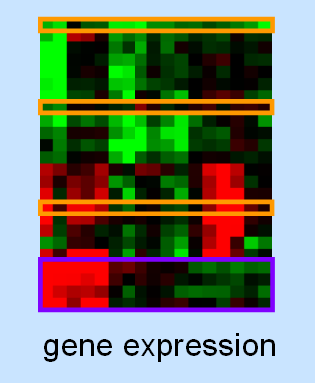
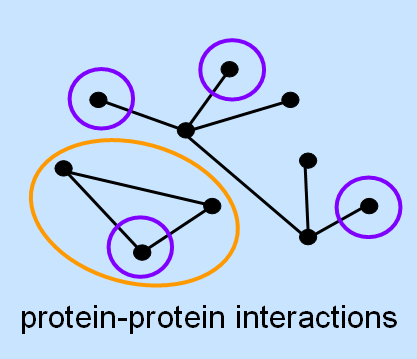
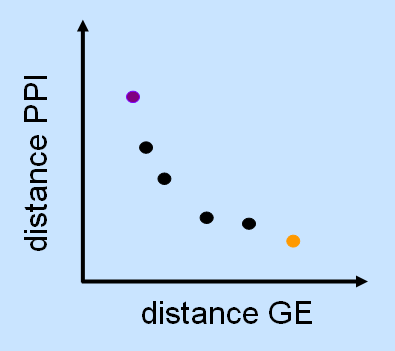
A central goal of postgenomic research is to assign a function to every predicted gene. Because genes often cooperate in order to establish and regulate cellular events, the examination of a gene has also included the search for at least a few interacting genes. This requires a strong hypothesis about possible interactions partners, which has often been derived from what was known about the gene or protein beforehand. Many times, though, this prior knowledge has either been completely lacking, biased towards favored concepts, or only partial due to the theoretically vast interaction space. With the advent of high-throughput technology and robotics in biological research, it has become possible to study gene function on a global scale, monitoring entire genomes and proteomes at once. These systematic approaches aim at considering all possible dependencies between genes or their products, thereby exploring the interaction space at a systems scale.
In this project, the goal is to develop methods for the identification of modules of genes or other entities from high-throughput data. On the one hand, we focus on advanced biclustering techniques that allow to find subgroups of genes that show the same response under a subset of conditions; in comparison to standard clustering methods, this approach often better reflects biological reality as genes can be related to multiple, distinct pathways. On the other hand, we develop concepts for the integration of multiple data types based on multiobjective models.
Own resources of the professorship.